Jupyter Notebook Server
This project uses the SecML library, which hasn't been updated since 2023. It no longer works in Google Colab. Instead, you need to use a local virtual machine running an older Jupyter Notebook.See project ML 107 to see how to download and run that machine.
We need training and testing sets, as usual. We also need a validation set, used to verify the classifier performance during the attack.
import secml
random_state = 999
n_features = 2 # Number of features
n_samples = 300 # Number of samples
centers = [[-1, -1], [1, 1]] # Centers of the clusters
cluster_std = 0.9 # Standard deviation of the clusters
from secml.data.loader import CDLRandomBlobs
dataset = CDLRandomBlobs(n_features=n_features,
centers=centers,
cluster_std=cluster_std,
n_samples=n_samples,
random_state=random_state).load()
n_tr = 100 # Number of training set samples
n_val = 100 # Number of validation set samples
n_ts = 100 # Number of test set samples
# Split in training, validation and test
from secml.data.splitter import CTrainTestSplit
splitter = CTrainTestSplit(
train_size=n_tr + n_val, test_size=n_ts, random_state=random_state)
tr_val, ts = splitter.split(dataset)
splitter = CTrainTestSplit(
train_size=n_tr, test_size=n_val, random_state=random_state)
tr, val = splitter.split(dataset)
# Normalize the data
from secml.ml.features import CNormalizerMinMax
nmz = CNormalizerMinMax()
tr.X = nmz.fit_transform(tr.X)
val.X = nmz.transform(val.X)
ts.X = nmz.transform(ts.X)
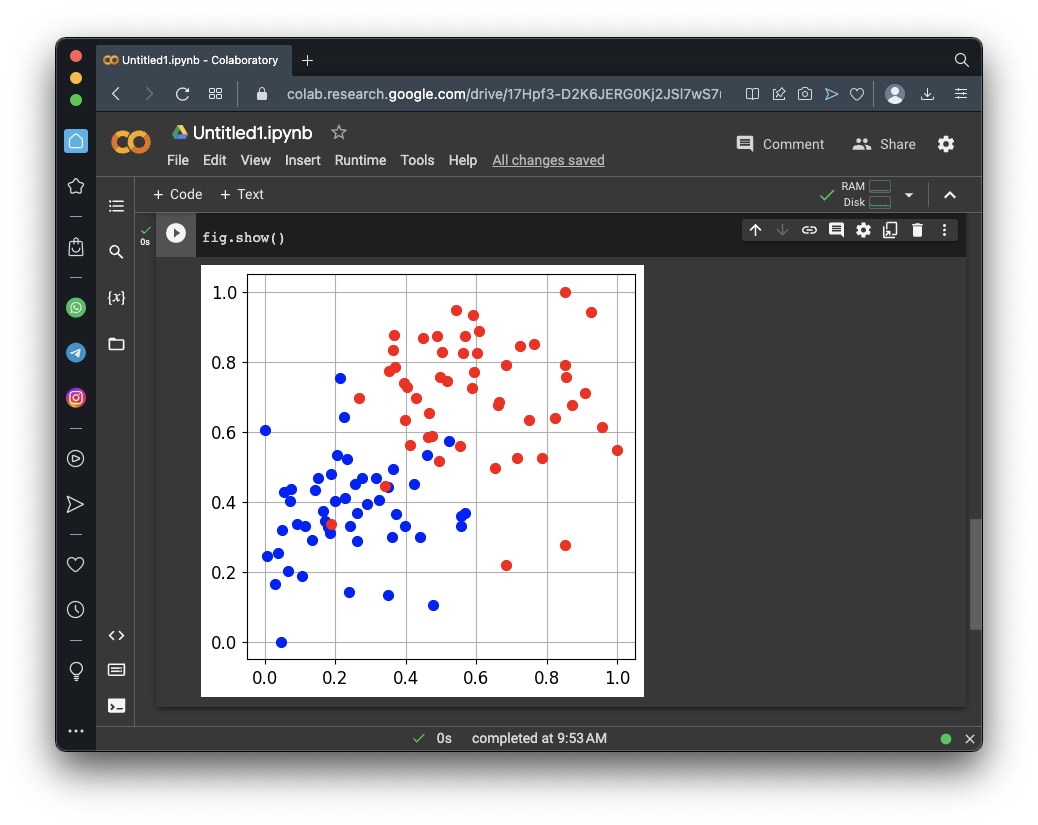
# Display the training set
from secml.figure import CFigure
# Only required for visualization in notebooks
%matplotlib inline
fig = CFigure(width=5, height=5)
# Convenience function for plotting a dataset
fig.sp.plot_ds(tr)
fig.show()
The task of this model is to sort the dots into their categories.
# Metric to use for training and performance evaluation
from secml.ml.peval.metrics import CMetricAccuracy
metric = CMetricAccuracy()
# Creation of the multiclass classifier
from secml.ml.classifiers import CClassifierSVM
from secml.ml.kernels import CKernelRBF
clf = CClassifierSVM(kernel=CKernelRBF(gamma=10), C=1)
# We can now fit the classifier
clf.fit(tr.X, tr.Y)
print("Training of classifier complete!")
# Compute predictions on a test set
y_pred = clf.predict(ts.X)
# Evaluate the accuracy of the classifier
acc = metric.performance_score(y_true=ts.Y, y_pred=y_pred)
print("Accuracy on test set: {:.2%}".format(acc))
fig = CFigure(width=5, height=5)
# Convenience function for plotting the decision function of a classifier
fig.sp.plot_decision_regions(clf, n_grid_points=200)
fig.sp.plot_ds(ts)
fig.sp.grid(grid_on=False)
fig.sp.title("Classification regions")
fig.sp.text(0.01, 0.01, "Accuracy on test set: {:.2%}".format(acc),
bbox=dict(facecolor='white'))
fig.show()
lb, ub = val.X.min(), val.X.max() # Bounds of the attack space. Can be set to `None` for unbounded
# Should be chosen depending on the optimization problem
solver_params = {
'eta': 0.05,
'eta_min': 0.05,
'eta_max': None,
'max_iter': 100,
'eps': 1e-6
}
from secml.adv.attacks import CAttackPoisoningSVM
pois_attack = CAttackPoisoningSVM(classifier=clf,
training_data=tr,
val=val,
lb=lb, ub=ub,
solver_params=solver_params,
random_seed=random_state)
# chose and set the initial poisoning sample features and label
xc = tr[0,:].X
yc = tr[0,:].Y
pois_attack.x0 = xc
pois_attack.xc = xc
pois_attack.yc = yc
print("Before poisoning: ({:.2f}, {:.2f}), label: {}".format(xc.ravel()[0].item(),
xc.ravel()[1].item(),
yc.item()))
from secml.figure import CFigure
# Only required for visualization in notebooks
%matplotlib inline
fig = CFigure(4,5)
grid_limits = [(lb - 0.1, ub + 0.1),
(lb - 0.1, ub + 0.1)]
fig.sp.plot_ds(tr)
# highlight the initial poisoning sample showing it as a star
fig.sp.plot_ds(tr[0,:], markers='*', markersize=16)
fig.sp.title('Attacker objective and gradients')
fig.sp.plot_fun(
func=pois_attack.objective_function,
grid_limits=grid_limits, plot_levels=False,
n_grid_points=10, colorbar=True)
# plot the box constraint
from secml.optim.constraints import CConstraintBox
box = fbox = CConstraintBox(lb=lb, ub=ub)
fig.sp.plot_constraint(box, grid_limits=grid_limits,
n_grid_points=10)
n_poisoning_points = 1 # Number of poisoning points to generate
pois_attack.n_points = n_poisoning_points
pois_y_pred, pois_scores, pois_ds, f_opt = pois_attack.run(ts.X, ts.Y)
fig.sp.plot_ds(pois_ds[0,:], markers='d', markersize=16)
xp = pois_ds[0,:].X
yp = pois_ds[0,:].Y
print("After poisoning: ({:.2f}, {:.2f}), label: {}".format(xp.ravel()[0].item(),
xp.ravel()[1].item(),
yp.item()))
fig.tight_layout()
fig.show()
The poisoning process moves the first point from the star, where it began, to the diamond shape above it, where the objective function is smaller.
n_poisoning_points = 1 # Number of poisoning points to generate
pois_attack.n_points = n_poisoning_points
# Run the poisoning attack
print("Attack started...")
pois_y_pred, pois_scores, pois_ds, f_opt = pois_attack.run(ts.X, ts.Y)
print("Attack complete!")
# Evaluate the accuracy of the original classifier
acc = metric.performance_score(y_true=ts.Y, y_pred=y_pred)
# Evaluate the accuracy after the poisoning attack
pois_acc = metric.performance_score(y_true=ts.Y, y_pred=pois_y_pred)
print("Original accuracy on test set: {:.2%}".format(acc))
print("Accuracy after attack on test set: {:.2%}".format(pois_acc))
# Training of the poisoned classifier
pois_clf = clf.deepcopy()
pois_tr = tr.append(pois_ds) # Join the training set with the poisoning points
pois_clf.fit(pois_tr.X, pois_tr.Y)
# Define common bounds for the subplots
min_limit = min(pois_tr.X.min(), ts.X.min())
max_limit = max(pois_tr.X.max(), ts.X.max())
grid_limits = [[min_limit, max_limit], [min_limit, max_limit]]
fig = CFigure(10, 10)
fig.subplot(2, 2, 1)
fig.sp.title("Original classifier (training set)")
fig.sp.plot_decision_regions(
clf, n_grid_points=200, grid_limits=grid_limits)
fig.sp.plot_ds(tr, markersize=5)
fig.sp.grid(grid_on=False)
fig.subplot(2, 2, 2)
fig.sp.title("Poisoned classifier (training set + poisoning points)")
fig.sp.plot_decision_regions(
pois_clf, n_grid_points=200, grid_limits=grid_limits)
fig.sp.plot_ds(tr, markersize=5)
fig.sp.plot_ds(pois_ds, markers='*', markersize=12)
fig.sp.grid(grid_on=False)
fig.subplot(2, 2, 3)
fig.sp.title("Original classifier (test set)")
fig.sp.plot_decision_regions(
clf, n_grid_points=200, grid_limits=grid_limits)
fig.sp.plot_ds(ts, markersize=5)
fig.sp.text(0.05, -0.25, "Accuracy on test set: {:.2%}".format(acc),
bbox=dict(facecolor='white'))
fig.sp.grid(grid_on=False)
fig.subplot(2, 2, 4)
fig.sp.title("Poisoned classifier (test set)")
fig.sp.plot_decision_regions(
pois_clf, n_grid_points=200, grid_limits=grid_limits)
fig.sp.plot_ds(ts, markersize=5)
fig.sp.text(0.05, -0.25, "Accuracy on test set: {:.2%}".format(pois_acc),
bbox=dict(facecolor='white'))
fig.sp.grid(grid_on=False)
fig.show()
On the right side, the top chart shows the data with a single poisining point (the star).
This point distorts the red-blue border at the top, near the added dot, but there's only one test data dot in that region, so it has only a minimal effect on the accuracy.
Flag ML 110.1: Poisoning More Points (10 pts)
Adjust the attack above to poison the first 20 dots in the training set.The flag is covered by a green rectangle in the image below.
Flag ML 110.2: Poisoning all the Points (5 pts)
Adjust the attack above to poison all 100 dots in the training set.The flag is covered by a green rectangle in the image below.
Flag ML 110.3: The Best Seed (15 pts)
Adjust the attack above to poison 5 dots.There are three blue poison dots and two red ones, shown as stars in the image below.
Examine the "Preparing a Dataset" code above. The seed for the pseudorandom number generator (random_state) was set to 999.In the "Visualizing Gradient Descent" code, that same seed is used again in the pois_attack definition.
Change the seed in the "Visualizing Gradient Descent" code to 0. This will choose different poison points, as shown below.
Try seed values from 0 to 5. Use the seed that causes the largest decrease in performance.The flag is covered by a green rectangle in the image below.
Posted 5-4-23
Jupyter server note added 5-15-25